|
Talairach
realignment warps an individual brain to a standard. Ideally, this
allows one to identify anatomical structures based on their absolute
Talairach coordinates. However, it warps the brain into a rather
unnatural flattened shape and the accuracy of the results is always
worth calling into question. All
examples are for the tutorial. Download
study1_norm+orig.BRIK and HEAD and brain+orig.BRIK and HEAD
from the analysis directory. Download
brain3d_cent+orig.BRIK and HEAD from the finished directory.
Note
that if you want to play with the talairached files, brain3d+tlrc
is available in the finished directory. You should end up with these
6 files to practice talairaching:
- study1_norm+orig.BRIK
and .HEAD,
- brain+orig.BRIK
and .HEAD,
- brain3d_cent+orig.BRIK
and .HEAD (the centered/aligned version)
How to
Start
Begin
with a 3d volume that has been preprocessed and centered. Go to
the directory containing that file and start afni:
>
afni
This
opens the afni graphical user interface (GUI):
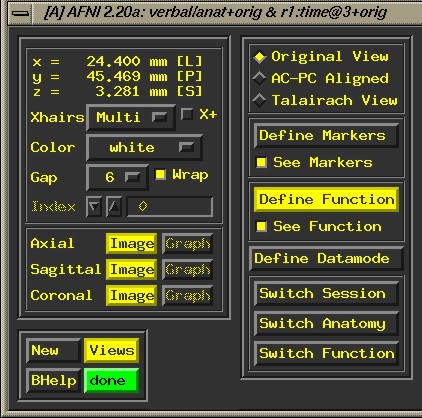
Choose
"Switch Anatomy" from the lower right corner, and select
the 3d volume that you wish to work with (choose "Set"
to select volume and close the popup menu).
On the left (middle box), you should select all three different
views to assist you in identifying the Talairach anchor points:
- Axial
"Image"
- Sagittal
"Image"
- Coronal
"Image"
Additionally:
- Make
sure "Original View" is selected in the upper right
- Select
"Define Markers" and the gui will be extended on the
right.
- Select
"Allow Edits" from the extended menu to the right so
you can pick the first set of anchor points for AC-PC alignment.
Anatomical
Anchors for AC-PC Alignment
- Consult
the following website for more advice:
http://afni.nimh.nih.gov/afni/
- Next
you must locate the following anchor points on the 3D brain image.
Once you have located an anchor point and placed your cross hairs
on it, select "Set" and go on to the next one. It is
preferable to locate points on the same plane, though it is not
always possible.
- Superior
Edge of Anterior Commisure: Click "AC superior edge".
Move up through the axials until you can't see the AC anymore.
Then go down one slice to where you can see it. This is the superior
edge. Click "set". In our tutorial data, the superior
edge of the AC is located on axial slice 122, coronal slice 113,
sagittal slice 61; x=4.950 mm L, y=-18.160 mm A, z=-5.571 mm I.
- Posterior
Margin of Anterior Commisure: Relying on the axial view, find
the image in which the AC is thickest. Find the back side of the
AC on this image. This is the posterior edge. In our tutorial
data, the posterior margin of the AC is located on
axial
slice 120, coronal slice 115, sagittal slice 61;
(x=4.950 mm L, y=-16.207 mm A, z=-7.524 mm I).
- Inferior
Edge of the Posterior Commisure: The PC is usually more difficult
to see than the AC. Using the axials and coronals, find the line
of white matter crossing between the two hemispheres above the
colliculi. Open the sagittal image to make sure you can get the
anterior and posterior commisures in the same sagittal plane.
For our tutorial data,
axial slice 113, coronal slice 140, sagittal slice 61;
x=4.950, y=8.207, z=-14.360.
- Two
Midsagittal Points:
Using the midsagittal image (the sagittal image that opens by
default in afni), try to find two points in the midcortical region,
preferably in the same plane as the AC and PC anchor points (which
are marked with little green squares as soon as you select "set"
for each anchor point). From the AC, fan out to a position anterior
and superior for the first point. From the PC, fan out to a position
posterior and superior for the second point.
- Select
"Quality Report". If you have done well, both angular
measures in the quality report will be less than 4%. If you have
not done well, you can go back to your anchor points and selectively
clear each one and try again. Keep trying until you get a good
quality report.
- Transform
Data. After a good quality report, select "Transform Data".
Additional
Anchors
- Now
select "AC-PC Aligned".
- Select
"Define Markers".
- When
the "Define Markers" interface is displayed, select,
"Allow Edits"
- Again,
make sure all three views of the brain are open.
- You
want to find cortex (choose views with whiter cortex not greyer
meninges dispayed).
- You
will see a new set of anchor points to identify and set:
- Most
Anterior Point: Use all three views to help find the most
forward point in the cortex. In our tutorial data, this is
on:
axial slice 79, coronal slice 22 and sagittal slice 107
(x=-12.000 mm R, y=-73.000 mm A, z= 9.000 mm S).
- Most
Posterior Point: Use the coronal view to help find this.
In our tutorial data, this is on:
axial slice 66, coronal slice 197 and sagittal slice 85
(x=10.000 mm L, y=102.000 mm P, z= -4.000 mm I).
- Most
Superior Point: Use the axial view to help find this.
In our tutorial data, this is on:
axial slice 144, coronal slice 118 and sagittal slice 90
(x=5.000 mm L, y=23.000 mm P, z= 74.000 mm S).
- Most
Inferior Point: Use the coronal view to help find this.
Ignore the cerebellum. This point will be the bottom edge
of a temporal lobe. In
our tutorial data, this is on:
axial slice 32, coronal slice 110 and sagittal slice 149
(x=-54.000 mm R, y=15.000 mm P, z= -38.000 mm I).
- Most
Left Point: This is actually the rightmost side of the
image. In our tutorial data, this is on:
axial slice 67, coronal slice 138 and sagittal slice 31
(x=64.000 mm L, y=43.000 mm P, z= -3.000 mm I).
- Most
Right Point: This is actually the leftmost side of the
image. In our tutorial data, this is on:
axial slice 101, coronal slice 124 and sagittal slice 164
(x=-69.000 mm R, y=29.000 mm P, z= 31.000 mm S).
- When
you have finished, run the "Quality Report". If there
are too many errors, afni will not let you continue and you will
need to clear suspect anchor points and try again.
- When
the data is satisfactory, select "Transform Data". This
creates a tlrc HEAD file.
Talairach
View
- Now
select "Talairach View".
- Select
"Define Datamode".
- On
the Define Datamode interface, select "Warp Anat on Demend"
- Also
select: Anat resam mode "Li" (Linear anatomical resampling)
- Select:
Write "Anat". This will create a BRIK file with the
same name as your original 3d volume, but it will replace the
"orig" extension with "tlrc". You will only
see this additional extension if you look at a listing of your
files in the shell. Afni will not display a file with this extension
in its popup menus. Rather, it will allow you to select the original
file AND "Talairach View" to display the 3d volume with
Talairach coordinates.
Overlaying
Functionals
- *You
may need to close and reopen afni so that it can refresh its information
about available files. Afni seems to have real difficulties with
refreshing.
- At
the command line:
>adwarp -apar anat+tlrc -dpar func+orig
will write out the dataset func+tlrc. Substitute the names of
appropriate anatomical and functional datasets. You should also
be able to choose "Define Datamode" in the gui, and
then Write Many from the lower right panel.
- Select
one or more functional image briks from the popup menus (these
already should be preprocessed, centered and analyzed) and warp
these into Talairach views as well.
- After
you create the functional warping, you can select "Switch
Function" to view a functional image overlaid on the anatomy
(As long as you have selected "Warp function on demend").
Note that by choosing the 3D volume in Talairach View, you are,
in effect, 'demanding' that the functional image be warped to
it.
You
will find that your talairached files are larger than the originals,
and that they are resliced into 1 mm isotropic voxels (this is what
makes them bigger). You will also find that all talairached files
are the same size as each other (because they all have the same
voxel size).
Skull
Stripping
Once
you have talairached the high resolution images, you can strip off
the skull:
3dIntacranial -prefix output -anat input
>3dIntracranial
-prefix brain_stripped -anat brain+tlrc
You
can apply skull stripping to either the 2D or 3D high resolution
talairached images (e.g., brain or brain3d). After brain stripping,
you might want to go to Define Datamodes->Plugins->Render.
Render will allow you to see a volume rendering of the head. You
can try it on both the stripped and unstripped head. The
render plugin will allow you to create a movie frame by frame.
For
more interesting things you can do with a talairached image, see
information on the built in talairach
atlas and masking.
|