|
|
|
These are high resolution (256x256 output) *.MR files. There are generally 2 series: series 2 with ~ 17 images; series 4 with ~124 images. The latter is used to make 3D volumes. Examples here are for the tutorial data. To preprocess anatomical images, you'll need several files from the Afni Download page (AnatomyOrig.tar.gz, the Scripts directory, and, if you don't have it, the rename_anat script from the Recon.tar.gz package). Alternatively, if you'd like a copy of the preprocessed anatomical images, so that you can work on centering and aligning the 3D, then you'll want the files from the Analysis area. If you'd like to work with the finished files (3D is already centered and realigned) then you'll want files from the Finished area. Now gunzip and untar the AnatomyOrig.tar.gz file. This will create a new Anatomy subdirectory. (If you aren't working with the tutorial data, then you should make an Anatomy directory and copy *.MR files into it.) >gunzip AnatomyOrig.tar.gz >tar xvf AnatomyOrig.tar Series 2 Images "2D Anatomical" Rename MR files: >cd
Anatomy Note that this process will report not being able to find a lot of images. This is because it is set up to rename 99 images and there are only 17 images in Series 2. Data collected after September, 2002? Check out the new rename_anat2 or rename_anat3. Make a "2D" Structural brick: Move up to the parent directory so that the BRIK and HEAD file will be placed here instead of in the Anatomy directory: >cd .. >to3d -anat -prefix brain Anatomy/E21670S2* Data Collected after September, 2002? A new header size and new naming strategy necessitate some changes in protocol. See Scanner Updates: Header Size for more details. Note: A feature as of late 2001 allows to3d to automatically read the headers on some anatomy files. In this case, to3d will make the brik without bringing up the interface. If there is a problem with the brik or you don't like this (and want to read Bob's poetry), then you can force to3d to bring up the interface by adding -nosave to the command, e.g., >to3d -anat -prefix brain -nosave Anatomy/E21670S2* In the to3d interface, should you choose or need to bring it up, set the following: x
orientation Right to Left Series 4 (spgr) Images "3D Anatomical" Use the series with 100+ images (usually series 4, but sometimes series 3 or 5) to make a 3D volume (E21670S4 will be a series with 124 slices). Don't bother to create a separate subdirectory. Run the Rename procedure and the to3d procedure as described above. Use an appropriate prefix, like brain3d: >cd Anatomy >rename_anat [at the prompt, enter the exam and series number, e.g., E21670S4] >to3d -anat -prefix brain3d seqplus ’Anatomy/E21670S4*’ to3d is the command. -anat is a flag telling afni that this is an anatomical file. -prefix is a flag that is followed with the prefix that you want to be assigned to the output file. seqplus="sequential in the plus direction". Finally, list the path to the files to be used in the BRIK. In the to3d window, make the following settings (and keep careful notes, it is very confusing): Choose
square pixels, slice thickness, z: 1.5 (use square
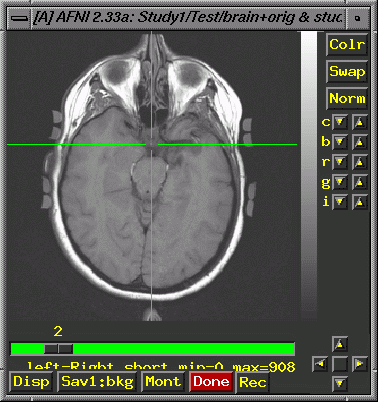
or irregular radio buttons to set). Field of view: 250 Centering Save Data, Quit. Unfortunately, when you display functional images overlaid on the 3D (Series 4) anatomy, they (the functionals) are likely to be noticeably off in terms of location on the anatomy. This is typical, and indicates that you must go beyond the centering (described above) and align the image manually to improve the overlay for Series 4. Keep in mind that here at the U of A, Series 2 (the 2D) is taken in the same plane as the functionals, and overlays of the functionals on the 2D are correct. We must, therefore, move the center of the Series 4 (3D) image to line up with the 2D image. Open both images (the 2D and the 3D) in Afni (use the "new" button in the lower left corner to get a 2nd set of Afni windows). Display activations (Define Function. This brings up additional widgets, a correlation bar and color bar. Select 4 colors, change orange to red and "set" the color change. If you leave the correlation slider at r=.5000, you'll rarely see activation, so check to make sure you have lowered it to 0. Click "See Function". You probably want to display a functional analysis with some reasonable p-value, which assumes you have done the analysis and have some reasonable file to display. Check under "Switch Function"). Find a distinct point that you can see on both images. The most superior point of the infundibular stalk (that little white dot that shows up on axial slices, just posterior to the optic chiasm; see the crosshairs in the image below) is a good choice. Start with the Series 2 image (the 2D image, brain+orig.BRIK) because it will have fewer slices, and so you are more likely to be able to find the same point in the 3D, than vice versa):
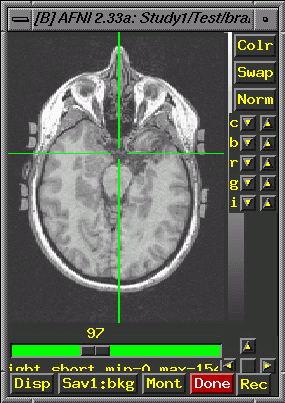
Put the crosshairs on the point and note the coordinates in the upper left corner of the afni window (e.g., -0.430 R, -24.492 A, -36.000 I; these values come from the tutorial data set): Now, in the 3D image, try to find the same anatomical landmark in a slice that is as similar to the 2D slice as you can manage. Consider the image below. The infundibular stalk is visible, though this is not the most superior point of the stalk in the 3D. Approximately equivalent amounts of the cerebellum show in both images. The lateral ventricles are not showing on either image. Look carefully at detailed anatomical features in the 3D slices near your landmark and choose your final slice based on the totality of shared features. Keep in mind that this is an art, and that your match will only be close, not exact.
In some cases, you may need to use 3drotate to rotate one image to be closer to the other (but you should not need to do this for the tutorial). Once you have found the same anatomical landmark on the most similar slice possible, note the coordinates (again, these values are from the tutorial set): 6.45 L, -24.996 A, -29.985 I. Calculate the differences between the location of the anatomical landmark in the 2D and 3D images:
Now determine the direction in which you'd have to move the center point of the 3D image to align it with the 2D image. For our tutorial image, we need to make the following changes: 6.88 R, 0.504 P, 6.015 I. This should be relatively straightforward from an examination of the landmark coordinates for the two images and the differences between them. Finally, make appropriate additions and subtractions from the current center points (see Centering above) For
the tutorial data: To make the actual changes as calculated, use 3drefit. Here is the command you'll want to enter for the tutorial data: >3drefit -xorigin 128.0077 -yorigin 118.2967 -zorigin 89.57 brain3d+orig.BRIK Sometimes this takes a few tries to get it right. You should end up being able to move the crosshairs around on the two images and get similar xyz coordinates. |